-Search query
-Search result
Showing all 31 items for (author: wu & xw)

EMDB-35929: 
Cryo-EM structure of Ufd4 in complex with K29/48 triUb
Method: single particle / : Ai HS, Mao JX, Wu XW, Pan M, Liu L

EMDB-35931: 
cryo-EM structures of Ufd4 in complex with Ubc4-Ub
Method: single particle / : Ai HS, Mao JX, Wu XW, Cai HY, Pan M, Liu L

EMDB-36300: 
Cryo-EM structure of compound 9n bound ketone body receptor HCAR2-Gi signaling complex
Method: single particle / : Zhao C, Tian XW, Liu Y, Cheng L, Yan W, Shao ZH

EMDB-36312: 
Cryo-EM structure of compound 9n and niacin bound ketone body receptor HCAR2-Gi signaling complex
Method: single particle / : Zhao C, Tian XW, Liu Y, Cheng L, Yan W, Shao ZH

EMDB-36317: 
Cryo-EM structure of niacin bound ketone body receptor HCAR2-Gi signaling complex
Method: single particle / : Zhao C, Tian XW, Liu Y, Cheng L, Yan W, Shao ZH

EMDB-36318: 
Cryo-EM structure of MMF bound ketone body receptor HCAR2-Gi signaling complex
Method: single particle / : Zhao C, Tian XW, Liu Y, Cheng L, Yan W, Shao ZH

EMDB-36490: 
Cryo-EM map of human receptor R2
Method: single particle / : Zhao C, Tian XW, Liu Y, Cheng L, Yan W, Shao ZH

EMDB-36491: 
Cryo-EM map of human Gi heterotrimer
Method: single particle / : Zhao C, Tian XW, Liu Y, Cheng L, Yan W, Shao ZH

EMDB-36492: 
Cryo-EM map of human receptor R2
Method: single particle / : Zhao C, Tian XW, Liu Y, Cheng L, Yan W, Shao ZH

EMDB-36493: 
Cryo-EM map of human Gi heterotrimer
Method: single particle / : Zhao C, Tian XW, Liu Y, Cheng L, Yan W, Shao ZH

EMDB-36494: 
Cryo-EM map of human receptor R2
Method: single particle / : Zhao C, Tian XW, Liu Y, Cheng L, Yan W, Shao ZH

EMDB-36495: 
Cryo-EM map of human Gi heterotrimer
Method: single particle / : Zhao C, Tian XW, Liu Y, Cheng L, Yan W, Shao ZH

EMDB-36496: 
Cryo-EM map of human receptor R2
Method: single particle / : Zhao C, Tian XW, Liu Y, Cheng L, Yan W, Shao ZH

EMDB-36497: 
Cryo-EM map of human Gi heterotrimer
Method: single particle / : Zhao C, Tian XW, Liu Y, Cheng L, Yan W, Shao ZH

EMDB-36505: 
Cryo-EM map of human receptor R2 in complex with Gi protein
Method: single particle / : Zhao C, Tian XW, Liu Y, Cheng L, Yan W, Shao ZH

EMDB-36506: 
Cryo-EM map of human receptor R2 in complex with Gi protein
Method: single particle / : Zhao C, Tian XW, Liu Y, Cheng L, Yan W, Shao ZH

EMDB-36507: 
Cryo-EM map of human receptor R2 in complex with Gi protein
Method: single particle / : Zhao C, Tian XW, Liu Y, Cheng L, Yan W, Shao ZH

EMDB-36508: 
Cryo-EM map of human receptor R2 in complex with Gi protein
Method: single particle / : Zhao C, Tian XW, Liu Y, Cheng L, Yan W, Shao ZH
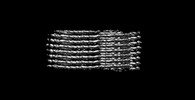
EMDB-40305: 
Cryo-EM structure of insulin amyloid-like fibril that is composed of two antiparallel protofilaments
Method: single particle / : Wang LW, Hall C, Uchikawa E, Chen DL, Choi E, Zhang XW, Bai XC

EMDB-23919: 
Cryo-EM structure of 2:2 c-MET/HGF holo-complex
Method: single particle / : Uchikawa E, Chen ZM, Xiao GY, Zhang XW, Bai XC

EMDB-23920: 
Cryo-EM structure of 1:1 c-MET I/HGF I complex after focused 3D refinement of holo-complex
Method: single particle / : Uchikawa E, Chen ZM, Xiao GY, Zhang XW, Bai XC

EMDB-23921: 
Cryo-EM map of the c-MET II/HGF I/HGF II (K4 and SPH) sub-complex
Method: single particle / : Uchikawa E, Chen ZM, Xiao GY, Zhang XW, Bai XC

EMDB-23922: 
Cryo-EM structure of the c-MET II/HGF I complex bound with HGF II in a rigid conformation
Method: single particle / : Uchikawa E, Chen ZM, Xiao GY, Zhang XW, Bai XC

EMDB-23923: 
Cryo-EM structure of 2:2 c-MET/NK1 complex
Method: single particle / : Uchikawa E, Chen ZM, Xiao GY, Zhang XW, Bai XC

EMDB-20572: 
Cryo-EM structure of extracellular dimeric complex of RET/GFRAL/GDF15
Method: single particle / : Li J, Shang GJ, Chen YJ, Brautigam CA, Liou J, Zhang XW, Bai XC

EMDB-20573: 
Cryo-EM structure of RET/GFRAL/GDF15 extracellular complex. The 3D refinement was focused on one of two halves with C1 symmetry applied.
Method: single particle / : Li J, Shang GJ, Zhang XW, Bai XC

EMDB-20575: 
Cryo-EM structure of RET/GFRa1/GDNF extracellular complex
Method: single particle / : Li J, Shang GJ, Chen YJ, Brautigam CA, Liou J, Zhang XW, Bai XC

EMDB-20576: 
Cryo-EM structure of RET/GFRa2/NRTN extracellular complex. The 3D refinement was applied with C2 symmetry.
Method: single particle / : Li J, Shang GJ, Chen YJ, Brautigam CA, Liou J, Zhang XW, Bai XC

EMDB-20578: 
Cryo-EM structure of RET/GFRa2/NRTN extracellular complex in the tetrameric form
Method: single particle / : Li J, Shang GJ, Chen YJ, Brautigam CA, Liou J, Zhang XW, Bai XC

EMDB-20579: 
Cryo-EM structure of RET/GFRa3/ARTN extracellular complex. The 3D refinement was applied with C2 symmetry.
Method: single particle / : Li J, Shang GJ, Chen YJ, Brautigam CA, Liou J, Zhang XW, Bai XC
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
